The CellCognition Project
Our mission is the development of a fast image analysis framework for fluorescence time-lapse microscopy in the field of bio-image informatics.
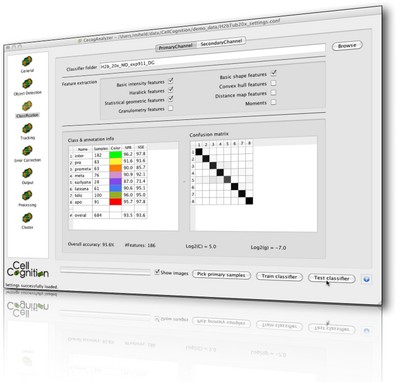
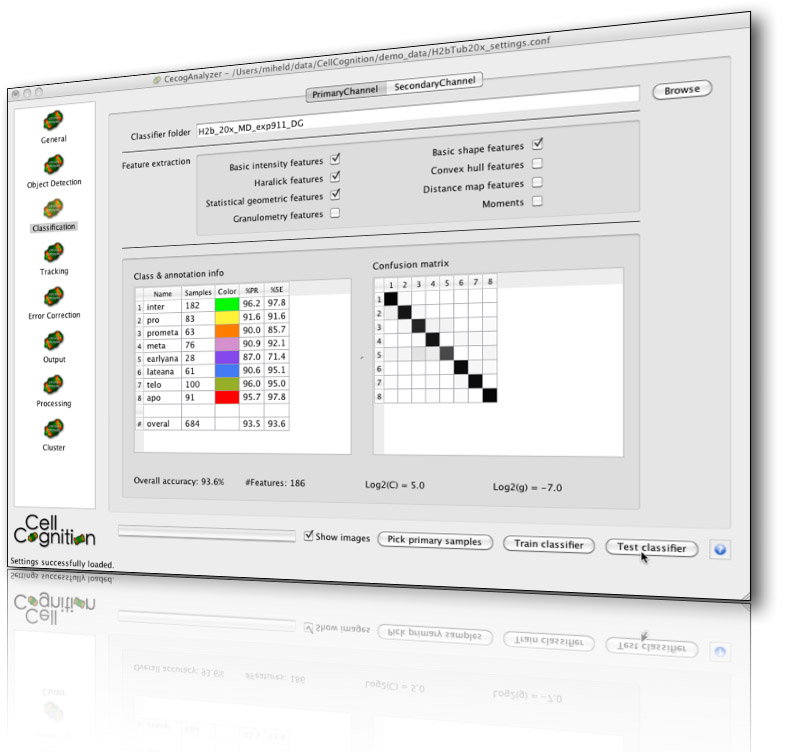
CellCognition is a computational framework dedicated to the automatic analysis of live cell imaging data in the context of High-Content Screening (HCS). It contains algorithms for segmentation of cells and cellular compartments based on various fluorescent markers, features to describe cellular morphology by both texture and shape, tools for visualizing and annotating the phenotypes, classification, tracking and error correction. Events such as mitosis can be automatically identified and aligned to study the temporal kinetics of various cellular processes during cell cycle. CellCognition can be used by novices in the field of image analysis and is applicable to hundreds of thousands of images by parallelization on compute clusters with minimal effort. The tool has been successfully applied to quantitative phenotypic profiling of cell division, yet machine learning enables CellCognition to be used for the analysis of other dynamic processes.
CellCognition is a common software project between the IMBA Vienna, Institut Curie Paris, EMBL Heidelberg, and ETH Zurich. With this framework we developed the CecogAnalyzer - a graphical user-interface for the parametrization and execution of our analysis workflow on a local machine and the submission to a computational cluster. We have further developed CellCognition Explorer, a software that offers a user interface optimized for interactive learning.